200以上 miniseq cluster density 133488-Miseq low cluster density
See performance parameters and specifications for the MiniSeq System The small footprint allows it to fit seamlessly into laboratoriesThis video provides an overview of overclustering and how it can impact your sequencing data An Illumina Field Applications Scientist shows you how to use SIf someone already owns a MiniSeq, convincing them to buy 2x250 kits is probably easier than convincing them to upgrade to (or expand with) a MiSeq A 2x250 mode on MiniSeq would be projected to yield about 105 gigabases of data, which is actually better than the MiSeq v2 2x250 mode (again, because of higher cluster density)

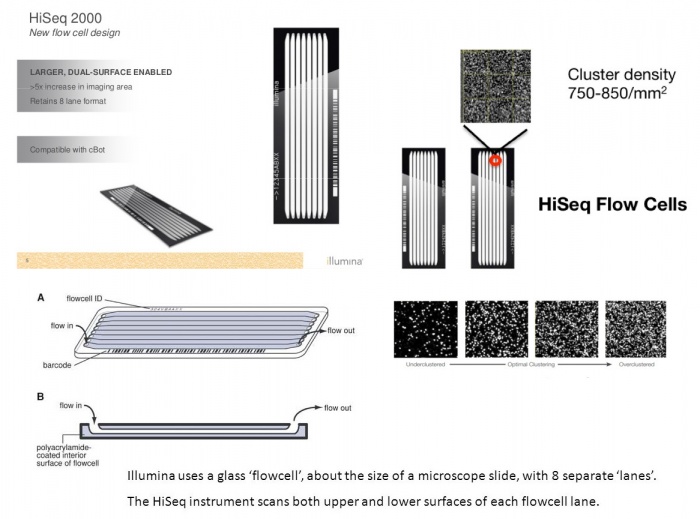
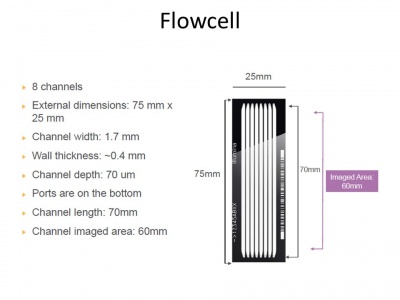
Next Generation Sequencing
Miseq low cluster density
Miseq low cluster density-Recorded Webinar (January 21) Illumina nextgeneration sequencing technology allows for massive parallel sequencing Illumina Technical Support invites you to join us for a presentation and discussion on sample preparation, cluster generation and sequencing by synthesis chemistry This webinar is targeted to those new to nextgeneration sequencing and will discuss the following topics Illumina library construction, clusterオーバークラスターしているフローセルのCluster densityは 実際よりも低い数値となる 2) サムネイル画像の確認 推奨最大cluster density/mm2 • MiSeq V2試薬キット 0965k/mm2 V3試薬キット k/mm2 •HiSeq00 k/mm2 •GAIIx k/mm2 原因 オーバークラスター



Callooooooom Necessary Miseq Run Sorry Nanopore Feeling Like I Ve Been Cheating Nanopolish
Achieving optimal cluster density is critical to highquality sequencing on MiniSeq, MiSeq, NextSeq, and HiSeq 2500 Systems Read more about how to achieve consistent cluster density in order to streamline data outputs Read Bulletin Product Highlights MiSeq Reagent Kits v3 enable the highest output of all MiSeq kits The v3 kits retain the same prefilled, readytouse reagentCluster density and flow cell loading recommendations in Table 1 assume you have a library that's sufficiently diverse If your library is not diverse or sufficiently balanced reduce the library loading amounts recommended in Table 1 by at least 10 % The exact reduction needed depends on several factors such as your library's insert size and whether the first bases of read 1 are · According to Illumina's specifications (Illumina 16b), the recommended cluster density for the mid‐output kit (300 cycles) on the MiniSeq is 170‐2 K/mm 2 Hence, we optimized cluster density by increasing the genetic diversity of the samples for sequencing runs B, C and D, by spiking in an additional Illumina library of genomic DNA from a marine sponge at a
Explore the Illumina workflow, including sequencing by synthesis (SBS) technology, in 3dimensional detail Go from sample preparation, to cluster generationAchieving optimal cluster density is critical to highquality sequencing on MiniSeq™, MiSeq™, NextSeq™ 500/550, and HiSeq™ 2500 Systems On nonpatterned flow cells, cluster density has a significant impact on run performance, specifically data quality and total data outputOverview of how the MiniSeq works, its features, how dual indexing is performed, best practices, and introductory guidance on run analysis View Recorded Webinar Sequencing Optimizing Cluster Density Learn about tips for achieving optimal cluster density View Recorded Webinar × Windows 10 Upgrade Upgrade to MiniSeq Software Suite v2 The MiniSeq system upgrade is
Cluster densities between 0–965 k/mm2 that pass filtering Actual performance may vary based on sample ††The percentage of bases > Q30 is averaged across the entire run, not on a perread or percycle basis bp = base pairs, Mb = megabases, Gb = gigabases, M = millions The information regarding future instrument specifications is intended to outline general product direction and itAchieving optimal cluster density is critical to highquality sequencing on MiniSeq, MiSeq, NextSeq, and HiSeq 2500 Systems Read more about how to achieve consistent cluster density in order to streamline data outputs Read Bulletin Product Highlights NextSeq 500/550 v25 sequencing reagent kits deliver the power of highthroughput sequencing on a benchtop system** Install specifications based on Illumina PhiX control library at supported cluster densities ( k/mm 2 clusters passing filter for v2 chemistry and k/mm 2 clusters passing filter for v3 chemistry) Actual performance parameters may vary based on sample type, sample quality, and clusters passing filter



Nebnext Library Quant Kit For Illumina Bioke


Cluster Density Vs Pf On Hiscansq And Hiseq Seqanswers
MethylMiniSeq Sample Report ZYMO RESEARCH CORP Phone (949) Toll Free (8) 6 Fax (949) services@zymoresearchcom info@zymoresearchcom wwwzymoresearchcom Materials & Methods The samples were processed and analyzed using the Classic RRBS Service Genomewide bisulfite sequencing (Zymo Research, Irvine, CA) DNAMuch higher clustering densities are achieved than with prior patterned flowcells, What I'd really like to see is an update to the MiSeq or MiniSeq that brings 2x higher cluster density and enables more consistent loading/passing filter 50M (100M even better) clusters would be ideal for most of our applications Could this be possible with a patterned flow cell?Density (K/mm 2) The density of clusters on the flowcell (in thousands per mm 2) On MiniSeq, MiSeq, NextSeq and HiSeq 2500 this is an important metric to evaluate if the data are lowquality It should be assessed in tandem with %PF as the two together can diagnose problems with over or underloading your library On HiSeq 4000 and X density is a fixed value and diagnosing issues


Quantifying Illumina Next Gen Sequencing Libraries Using Digital Pcr On The Quantstudio 3d Youtube



Developmental Validation Of The Miseq Fgx Forensic Genomics System For Targeted Next Generation Sequencing In Forensic Dna Casework And Database Laboratories Forensic Science International Genetics
Cluster density is a critically important metric that influences run quality, reads passing filter, Q30 scores, and total data output While underclustering maintains highAchieving optimal cluster density is critical to highquality sequencing on MiniSeq™, MiSeq™, NextSeq™ 500/550, and HiSeq™ 2500 Systems On nonpatterned flow cells, cluster density has a significant impact on run performance, specifically data quality and total data output While underclustering can maintain high data quality, it results in lower data output AlternativelyTotal times include cluster generation, sequencing and base calling on a NextSeq 500 System Install specifications are based on Illumina PhiX control library at supported cluster densities (between 170 and 230 k/mm2 clusters passing filter Actual performance parameters may vary based on sample type, sample quality, and clusters passing filter The percentage of bases >


Bilgenur Baloglu Check Out My Poster On Metabarcoding With Nanopore Sequencing Find It At 49 By Scrolling One Page Left Thanks Olga Francino Max Krau5e And Laura Boykin For Your Insightful Questions I



Dna Next Generation Sequencer Miseq Fgx Illumina Inc Forensic Medicine
Enrichment and sequencing with the MiniSeq Rapid Reagent Kit offers several advantages, including • Sensitive detection of both DNA and RNAbased respiratory pathogens in a single assay in under 24 hours • Comprehensive genome coverage of critical viral pathogens, including SARSCoV2 and Influenza A virus •the common cold (viral rhinitis), sinusitis, pharyngitis, andFOR RESEARCH USE ONLY ILLUMINA PROPRIETARY Part # Rev H March 13 MiSeq® System User Guide · However, it is important to note that random sequencing error, clustering density, percentage of reads passing the filter, and the overall noise in the sequencing signal resulted in 26, 29, and 26 variant calls when comparing the MiSeq run at UCSD and the MiSeq run at Illumina for samples 112, 027, and 066, respectively The WGS results demonstrate the ability to


Nick Loman There Is A Definite Sweet Spot For Miseq Cluster Density Same Library Gc 65 Nextera Xt Wgs V2 Kits Http T Co Mtqo8yadx3



Coregenomics Nextseq 500 S New Chemistry Described
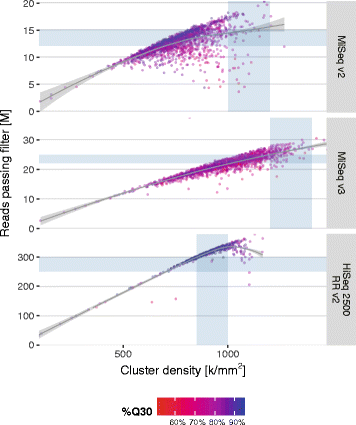
PDF In the last decades, methanogenic archaea have been increasingly investigated with regard to their impact on the environment, technological Find, read and cite all the research you needSee performance parameters and specifications for the MiniSeq System The small footprint allows it to fit seamlessly into laboratoriesOn the MiSeq sequencing platform, the optimal raw cluster density specifications for balanced libraries differ by reagent chemistry version • MiSeq v2 reagents accommodate an optimal raw cluster density of 1000–10 K/mm² • MiSeq v3 reagents accommodate an optimal raw cluster density of 10–1400 K/mm²



The Comparison Between Pgm And Miseq Download Table



1 Miseq Flip Ebook Pages 1 4 Anyflip Anyflip
Cluster Generation and Sequencing * Install specifications based on Illumina PhiX control library at supported cluster densities (between 129 and 165 k/mm 2 clusters passing filter) Actual performance parameters may vary based on sample type, sample quality, and clusters passing filter All MiniSeq kits are pairedend compatibleLocal Run Manager v2 is available for MiniSeq instruments using MiniSeq Control Software v2 or later, NextSeq 500/550 instruments using NextSeq Control Software v4 and later, and MiSeq instruments using MiSeq Control Software v3 Languages French, Italian, German, Spanish, Chinese (Simplified) min Optimal Cluster Density Best Practices This video discusses optimal cluster densityToday I put on a MiSeq run using Nextera XT library preparation to be run on a V3 600 cartridge After ~ cycles it has given me a low cluster density of 473K/mm2 The quality however is


The Quantitation Question How Does Accurate Library Quantitation Influence Sequencing Neb



Helix Bits And Codes Tile Shapes In Hiseq And Miseq
Consult the bulletin, Cluster density considerations when migrating Illumina libraries between sequencing platforms for additional information on these variations Additional considerations Insert sizes more than 550 bp are generally not supported on the MiniSeq, NextSeq 500/550, or HiSeq 3000/4000 platforms, and may require additional optimization steps Some applicationsAccording to Illumina's specifications (Illumina 16b), the recommended cluster density for the mid‐output kit (300 cycles) on the MiniSeq is 170‐2 K/mm 2 Hence, we optimized cluster density by increasing the genetic diversity of the samples for sequencing runs B, C and D, by spiking in an additional Illumina library of genomic DNA from a marine sponge at a ratio of 13Library input concentration also has an effect on cluster density and the concentration amount can be different in each lab The usual concentration for optimal cluster density is about 515 pM Library Nucleotide Diversity Library nucleotide diversity is another factor to consider when determining how balanced your library is for the nucleotides in order to achieve maximum



I Had A Metagenomic 16s Rrna Run On Miseq V3 Kit 2 X300 Cycles With Good Pf But Low Quality Q30 40 Kindly Advice What Could Get Wrong


New Thing Archive Seqanswers
· 129 According to Illumina's specifications (Illumina 16), the recommended cluster density for the 130 midoutput kit (300 cycles) on the MiniSeq is 1702 K/mm 2Page 54 Optimally, the cluster density should be 800K/mm² However, a range of 50K– 1300K/mm² is acceptable Clusters Passing Filter (%)—Shows the percentage of clusters passing filter based on the Illumina chastity filter, which measures quality This data appears only after cycle 25 NOTE The chastity of a base call is the ratio of the intensity of the greatest signal divided byYou may need to manually stop and restart RTA if it is stalled or if the network path is down For more information, see the bulletin How to restart RTA on MiSeq system Cluster density is a critically important metric that influences run quality, reads passing filter, Q30 scores, and total data output



A B Results Of 30 Bone Samples Run On Fgx Miseq Showing A Cluster Download Scientific Diagram



V4some 1tb Here We Come Enseqlopedia
** Install specifications based on Illumina PhiX control library at supported cluster densities ( k/mm 2 clusters passing filter for v2 chemistry and k/mm 2 clusters passing filter for v3 chemistry) Actual performance parameters may vary based on sample type, sample quality, and clusters passing filterThe MiniSeq and NextSeq 500/550 platforms require significantly lower cluster densities than the HiSeq and MiSeq platforms Therefore, it is important to redo cluster density optimization when migrating a library from MiSeq or HiSeq to MiniSeq or NextSeq 500/550 platform and vice versa To or from HiSeq 3000/4000 and NovaSeq 6000Training resources for the MiniSeq System This video discusses optimal cluster density and how over and underclustering can affect your sequencing data It also examines common clustering issues and ways to prevent them 10 min Data Analysis DATE/ LENGTH BaseSpace Sequence Hub Introduction and Analysis Overview Recorded Webinar (May ) llumina



Next Generation Sequencing



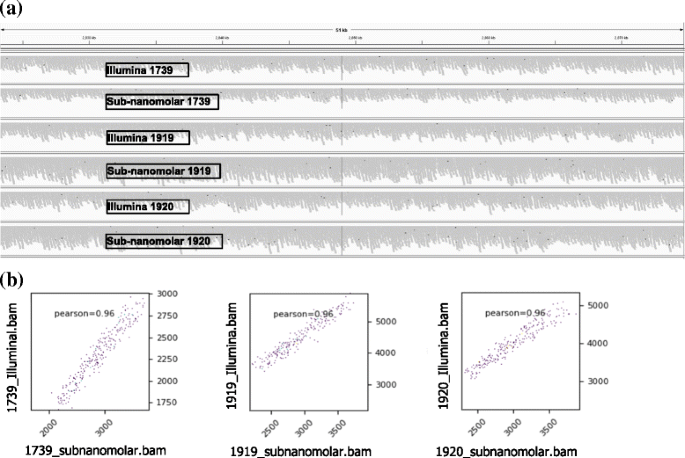
Details And Comparisons Of Illumina Protocol And Sub Nanomolar Download Scientific Diagram
MiniSeq MiniSeq High/Mid Output 170 –2 MiSeq MiSeq v3 10 –1400 MiSeq v2 1000 –10 NextSeq High/ Mid Output (v25 and v2) 170 2 ※ 上記推奨密度は塩基多様性に偏りのないライブラリーを使用した場合を想定The density of clusters on the flow cell (in thousands per mm 2) On MiniSeq, MiSeq, NextSeq and HiSeq 2500 this is an important metric to evaluate if the data are lowquality It should be assessed in tandem with %PF as the two together can diagnose problems with over or underloading your library On HiSeq 4000 and X density is a fixed value and diagnosing issues with library loadingOptimal Raw Cluster Density HiSeq™ High Output, v3 K/mm 2 High Output, v4 K/mm 2 Rapid Run, v2 K/mm 2 MiSeq™ v2 K/mm 2 v3 K/mm 2 MiniSeq™ Mid and High Output 1702 K/mm 2 NextSeq 500/550™ Mid and High Output 1702 K/mm 2



Smarter Stranded Total Rna Seq For Mammalian Samples



174 Questions With Answers In Illumina Sequencing Science Topic
· 1) Reduce your cluster density by 5080% to reduce overlapping clusters 2) Use a high amount of PhiX spike in (up to 50%) of the total library 3) Use custom primers with a random sequence to increase diversity Alternatively, intentionally concatamerize your amplicons and fragment them to increase base diversity at the start of your readsMiniSeq System features – Maximum Output 75 Gb (equivalent to 75 billion bases) within 24 hours – Data quality Quality Score > Q30 (999% accuracy) – Sequencing time when using the HighOutput Reagent Kits 1 x 50 bp in 7 hours (384 samples) 2 x 75 bp in 13 hours (23 samples) 2 x 150 bp in 24 hours (50 smallgenome sequencing samples, 28 targeted amplicon sequencingThe density of clusters for each tile (in thousands per mm2) The number of clusters for each tile (in millions) The estimated percentage of molecules in a cluster for which sequencing falls behind (phasing) or jumps ahead (prephasing) the current cycle within a read The percentage of reads from clusters in each tile that aligned to the PhiX



Miseq University Of Utah


Miseq Multiplex Fail Seqanswers
Local Run Manager is oninstrument analysis software integrated with the system control software Using Local Run Manager, you can record sample information and specify run parameters before starting the run The name assigned to the run appears on the control software run setup screen for faster run setupMiSeq Reagent Kits v2 How to Achieve More Consistent Cluster Density on Illumina Sequencing Platforms Achieving optimal cluster density is critical to highquality sequencing on MiniSeq, MiSeq, NextSeq, and HiSeq 2500 Systems Read more about how to achieve consistent cluster density in order to streamline data outputs



Miseq Imaging And Base Calling Pdf Free Download


Hiseq00 Next Level Hacking Hackteria Wiki



I Had A Metagenomic 16s Rrna Run On Miseq V3 Kit 2 X300 Cycles With Good Pf But Low Quality Q30 40 Kindly Advice What Could Get Wrong



Callooooooom Necessary Miseq Run Sorry Nanopore Feeling Like I Ve Been Cheating Nanopolish



Mads Albertsen على تويتر Hacking Our 2nd Miseq V3 Run But We Simply Couldn T Resist Might Fail Completely Though Http T Co Tmm8f4z5pe



Miniseq Twitter Search



Cluster Density Illumina Page 1 Line 17qq Com



Miseq System Focused Power For Targeted Gene And Small Genome Sequencing



Gdna Enrichment By A Transposase Based Technology For Ngs Analysis Of The Whole Sequence Of Brca1 Brca2 And 9 Genes Involved In Dna Damage Repair Protocol Translated To Dutch



Library Preparation And Miseq Sequencing For The Genotyping By Sequencing Of The Huntington Disease Htt Exon One Trinucleotide Repeat And The Quantification Of Somatic Mosaicism Protocol Exchange On Research Square



Miseq Possible Growth Potential Part 2 Enseqlopedia


Hiseq00 Next Level Hacking Hackteria Wiki



Scheellab First Miseq Run On Hcv Rna Interactions With Born2bewildtype



Miseq Possible Growth Potential Part 2 Enseqlopedia


Broad S Experience With The Miseq 300bp Reads Possible Seqanswers



Duplicates On Illumina



Next Generation Sequencing Ppt Download


Miseq Miseq System User Manual My Illumina



Diagnosing Suboptimal Clustering In Nonpatterned Flow Cells Youtube


Sequencing Quality Control Oxford Genomics Centre Oxford Genomics Centre



In Depth Comparative Analysis Of Illumina Miseq Run Metrics Development Of A Wet Lab Quality Assessment Tool Kastanis 19 Molecular Ecology Resources Wiley Online Library


New Thing Archive Seqanswers



Is My Hiseq Or Miseq Run Overclustered Illumina Video Youtube


Robust Sub Nanomolar Library Preparation For High Throughput Next Generation Sequencing Bmc Genomics Full Text


New Thing Archive Seqanswers
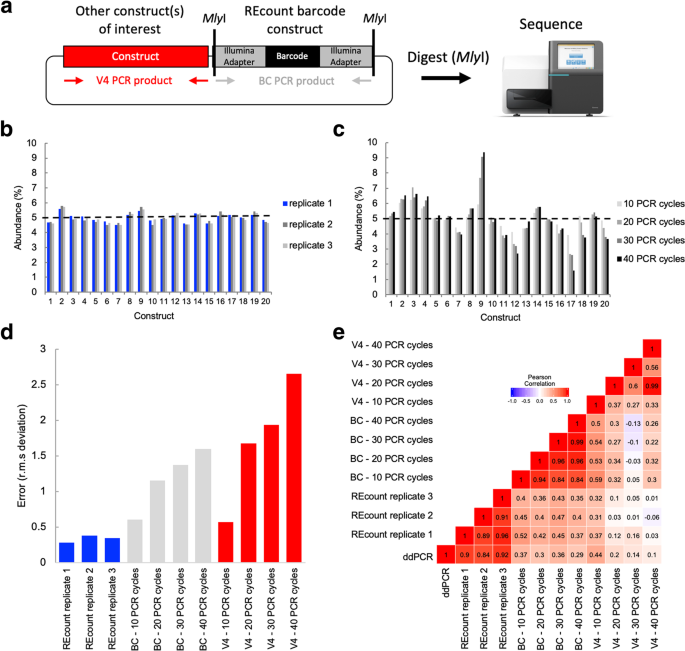


Measuring Sequencer Size Bias Using Recount A Novel Method For Highly Accurate Illumina Sequencing Based Quantification Genome Biology Full Text


2 7 Million Samples Genotyped For Hla By Next Generation Sequencing Lessons Learned Bmc Genomics Full Text



Illumina Dye Sequencing Wikipedia



Coregenomics Hacking Miseq Updated And Now Hacking Your Bioanalyser Too


Sequencing Quality Control Oxford Genomics Centre Oxford Genomics Centre



Differential Amplicon Size Effects On Clustering Across Illumina Instruments Seqanswers



Optimizing Miseq Library Cluster Density





Sequencing Quality Control Oxford Genomics Centre Oxford Genomics Centre



Building A Custom Large Scale Panel Of Novel Microhaplotypes For Forensic Identification Using Miseq And Ion S5 Massively Parallel Sequencing Systems Sciencedirect



C Sop 601 Operation Of The Illumina Miseq For Whole Genome Sequencing Wgs



I Had A Metagenomic 16s Rrna Run On Miseq V3 Kit 2 X300 Cycles With Good Pf But Low Quality Q30 40 Kindly Advice What Could Get Wrong



Box And Whisker Plots Summarizing Six Different Miseq Run Metrics Download Scientific Diagram



Next Generation Sequencing Tips N Tricks Part 4 Diagnostech


New Thing Archive Seqanswers



Developmental Validation Of The Miseq Fgx Forensic Genomics System For Targeted Next Generation Sequencing In Forensic Dna Casework And Database Laboratories Sciencedirect



Optimizing Cluster Density On Illumina Sequencing Systems Pdf Document


コメント
コメントを投稿